Search Count: 3,897
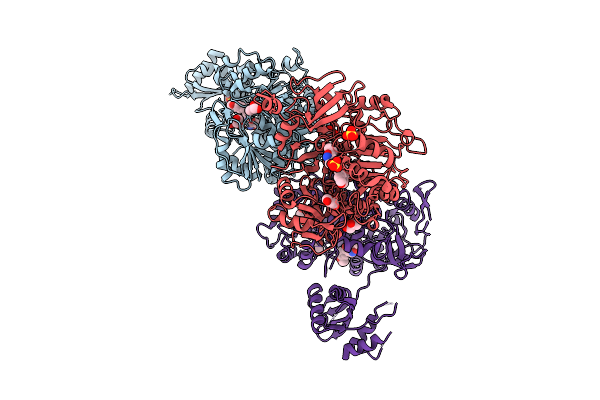 |
Crystal Structure Of Acetyl-Coa Synthetase From Cryptococcus Neoformans H99 In Complex With Inhibitor Hgn-1310 (Dd3-027)
Organism: Cryptococcus neoformans var. grubii
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LIGASE Ligands: GOL, SO4, CL, A1AV1 |
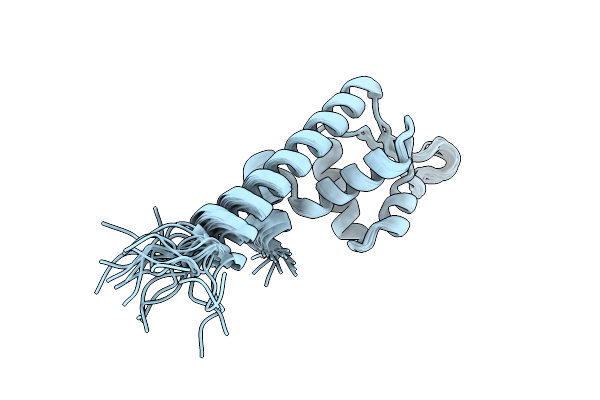 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
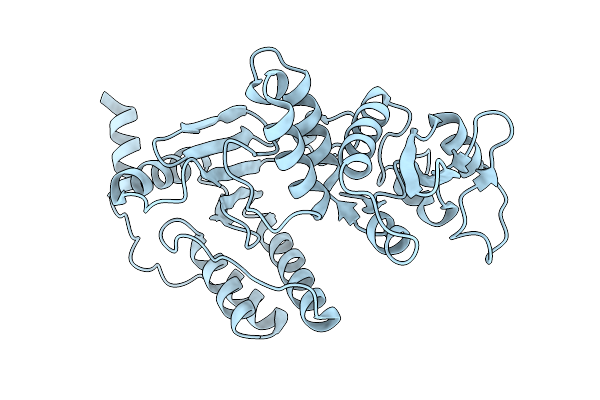 |
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN |
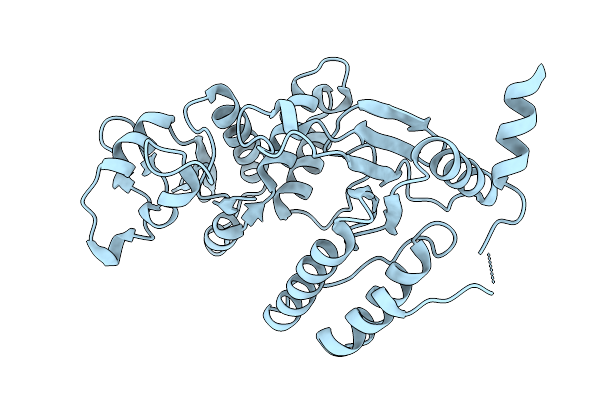 |
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN |
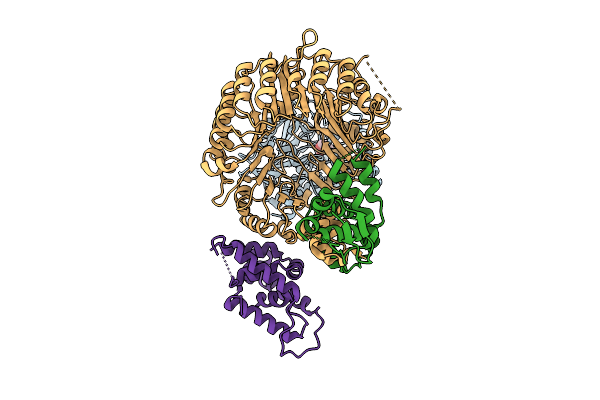 |
Strigolactone-Induced Ask1-Max2-Htl7-Smax1 Complex (Class 1) With Covalently Bound D-Ring
Organism: Arabidopsis thaliana, Striga hermonthica
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: GR2 |
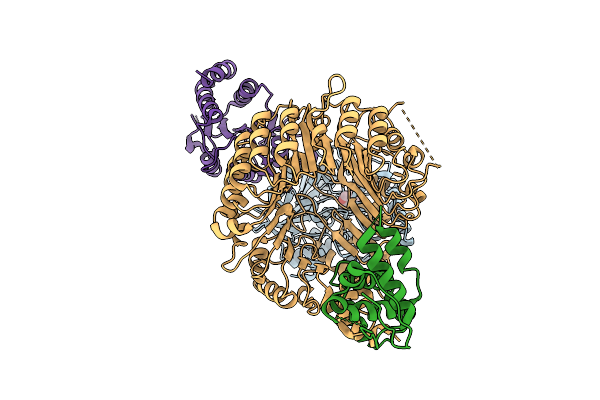 |
Strigolactone-Induced Ask1-Max2-Htl7-Smax1 Complex (Class 2) With Covalently Bound D-Ring
Organism: Arabidopsis thaliana, Striga hermonthica
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: GR2 |
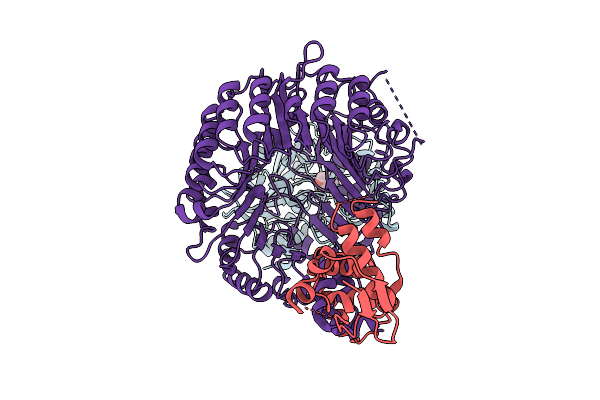 |
Strigolactone-Induced Ask1-Max2-Htl7-Smax1 Complex (Class 4) With Covalently Bound D-Ring
Organism: Arabidopsis thaliana, Striga hermonthica
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: GR2 |
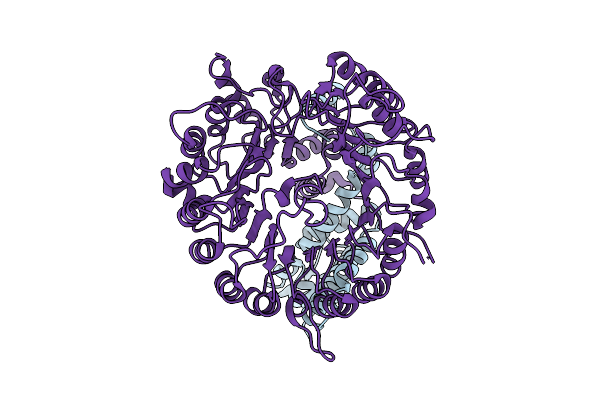 |
Organism: Arabidopsis thaliana, Striga hermonthica
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN |
 |
Strigolactone-Induced Ask1-Max2-Htl7-Smax1 Complex (Class 3) With Covalently Bound D-Ring
Organism: Arabidopsis thaliana, Striga hermonthica
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: GR2 |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LIGASE Ligands: LYS, GOL |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: SO4 |
 |
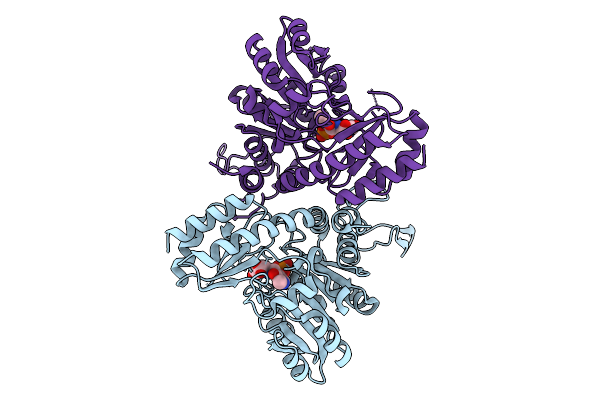
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: PEG |
 |
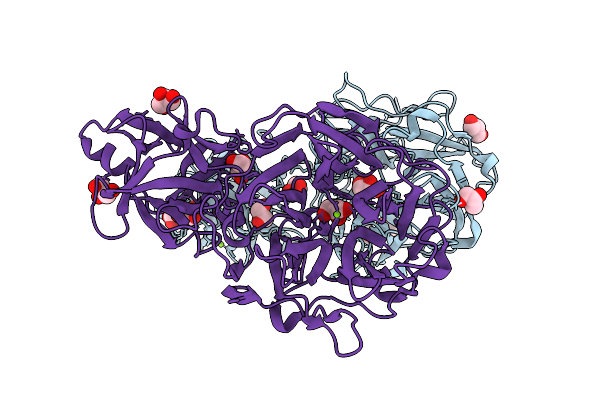
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: PGU |
 |
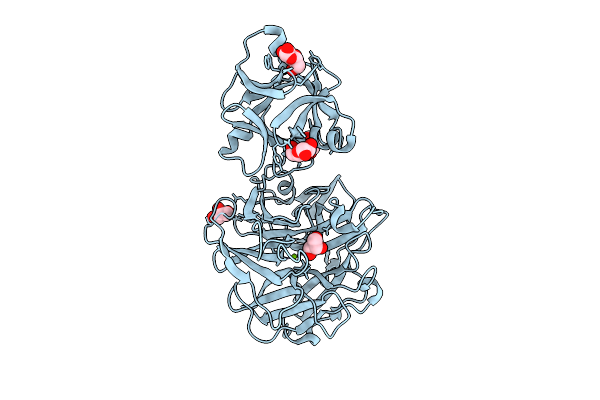
An Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: GOL, MG |
 |
An Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313 Bound To Arabinofuranose
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: AHR, GOL, MG |
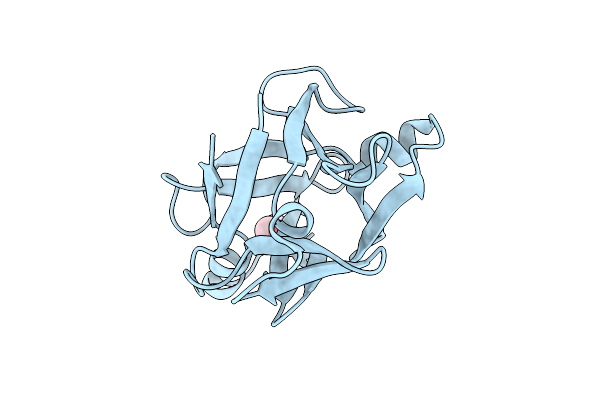 |
Cbm42 Domain Of Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: GOL |
 |
The Gh43 Domain Of An Alpha-L-Arabinofuranosidase (Atabf43C_Gh43) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: AHR, SO4 |
 |
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) Complex With Glcnac-1,6-Anhmurnac
Organism: Homo sapiens, Synthetic construct, Yokenella regensburgei, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
The Structure Of Cbm42 From Clostridium Thermocellum In Complex With Arabinofuranosyl Xylobiose (A3X) Substrate Molecules
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SUGAR BINDING PROTEIN Ligands: CL, GOL |

