Search Count: 32
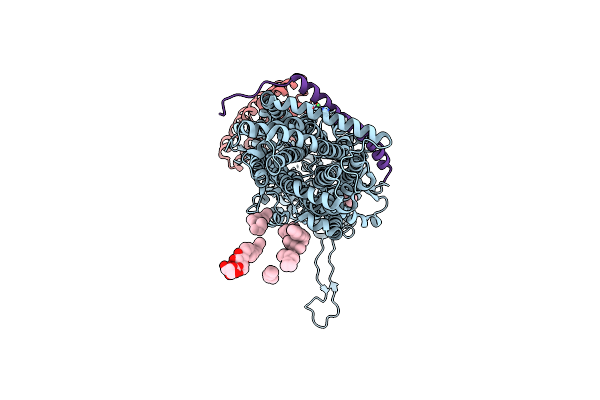 |
Organism: Escherichia coli k-12, Synthetic construct, escherichia coli str. k-12 substr. w3110
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-04-30 Classification: TRANSPORT PROTEIN Ligands: LMT, NI |
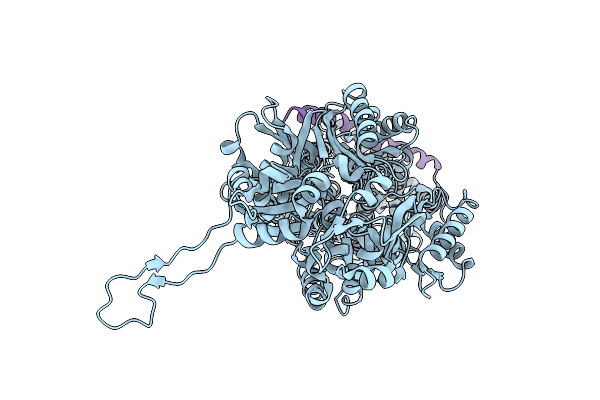 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2014-04-30 Classification: MEMBRANE PROTEIN |
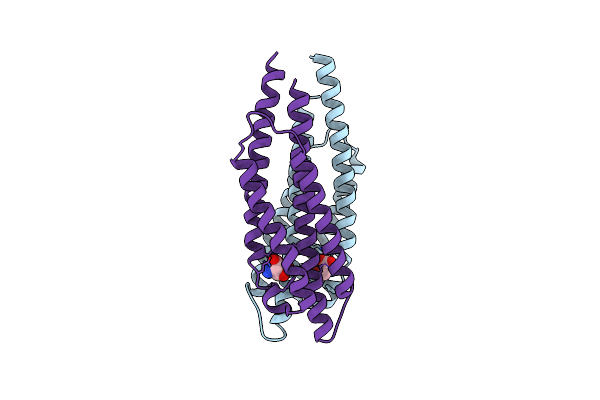 |
Structure Of The Ligand Binding Domain Of The Bacterial Serine Chemoreceptor Tsr With Ligand
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: SIGNALING PROTEIN Ligands: SER |
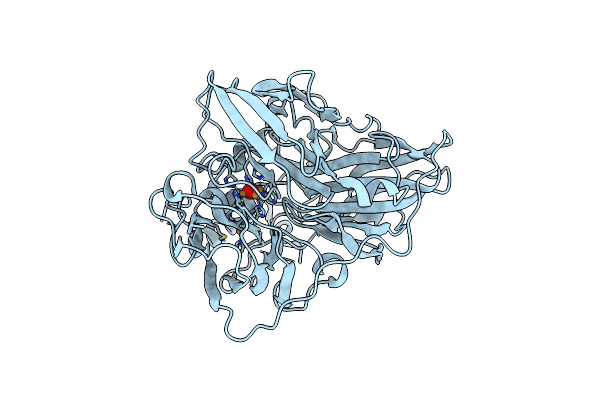 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-19 Classification: OXIDOREDUCTASE Ligands: C2O, CU |
 |
Structual Basis Of Azido-Tyrosine Recognition By Engineered Bacterial Tyrosyl-Trna Synthetase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-29 Classification: LIGASE Ligands: AZY |
 |
Crystal Structure Of Truncated Fimg (Fimgt) In Complex With The Donor Strand Peptide Of Fimf (Dsf)
Organism: Escherichia coli str. k12 substr., Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2008-03-04 Classification: STRUCTURAL PROTEIN/STRUCTURAL PROTEIN Ligands: CO |
 |
Crystal Structure Of Truncated Fimg (Fimgt) In Complex With The Donor Strand Peptide Of Fimf (Dsf)
Organism: Escherichia coli str. k12 substr., Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-03-04 Classification: STRUCTURAL PROTEIN/STRUCTURAL PROTEIN Ligands: YT3 |
 |
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2007-10-02 Classification: METAL BINDING PROTEIN Ligands: NO3, SO4, AG |
 |
Crystal Structure Of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase Complexed With A Magnesium Ion, Nadph And Fosmidomycin
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-06-19 Classification: OXIDOREDUCTASE Ligands: MG, FOM, NDP |
 |
Crystal Structure Of The Mutant N560D Of The Nuclease Domain Of Cole7 In Complex With Im7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2007-04-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, PO4 |
 |
Crystal Structure Of The Mutant H573A Of The Nuclease Domain Of Cole7 In Complex With Im7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2007-04-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN |
 |
Crystal Structure Of The Mutant N560A Of The Nuclease Domain Of Cole7 In Complex With Im7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-04-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, SO4 |
 |
Structure Of Acyl Carrier Protein Bound To Fabi, The Enoyl Reductase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-10-17 Classification: OXIDOREDUCTASE/BIOSYNTHETIC PROTEIN |
 |
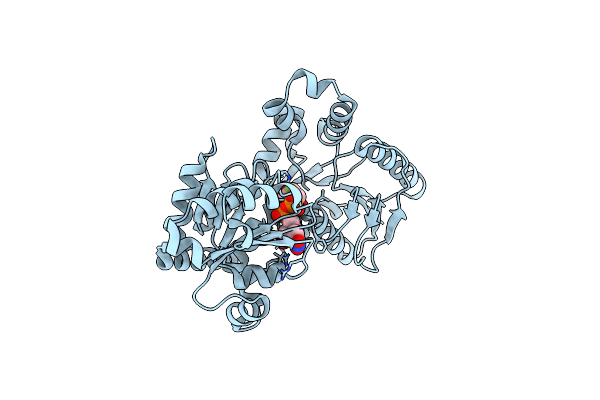
Crystal Structure Of Waag, A Glycosyltransferase Involved In Lipopolysaccharide Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-10-11 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Waag, A Glycosyltransferase Involved In Lipopolysaccharide Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-10-11 Classification: TRANSFERASE Ligands: U2F |
 |
Organism: Escherichia coli str. k12 substr.
Method: SOLUTION NMR Release Date: 2006-09-24 Classification: PROTEIN BINDING |
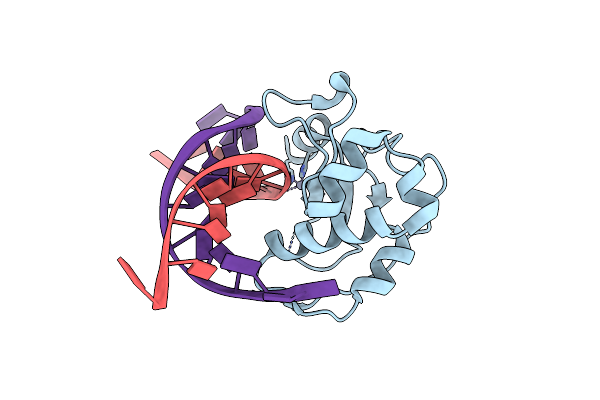 |
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-03-14 Classification: Hydrolase/DNA Ligands: ZN |
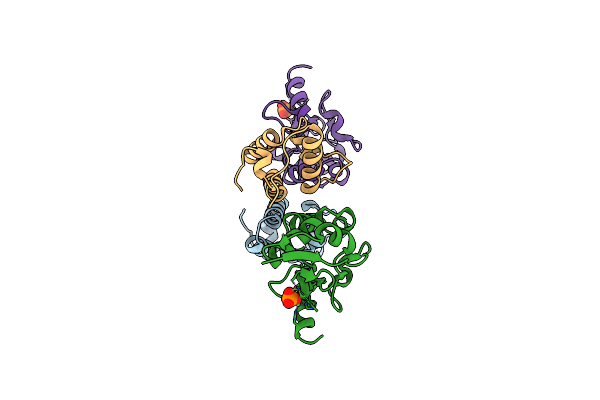 |
How A His-Metal Finger Endonuclease Cole7 Binds And Cleaves Dna With A Transition Metal Ion Cofactor
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-03-14 Classification: Hydrolase/PROTEIN BINDING Ligands: NI, PO4 |
 |
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal And C-Terminal Domain Of The Electron Transfer Catalyst Dsbd
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2005-07-12 Classification: OXIDOREDUCTASE |

