Search Count: 472
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: ISOMERASE Ligands: GDP, MG |
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: ISOMERASE |
 |
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: EDO, GLU, GLY, FES, FE2, GOL |
 |
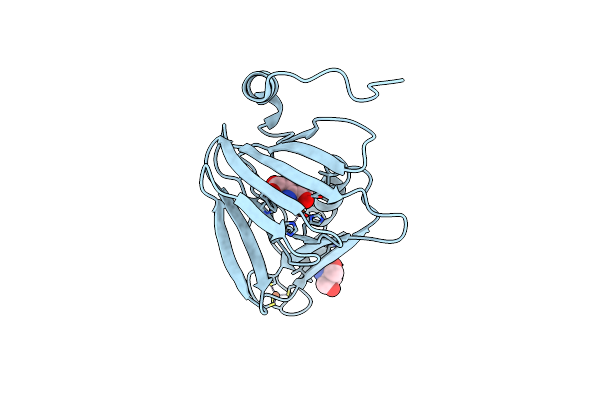
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - A Resting State Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE2, CL |
 |
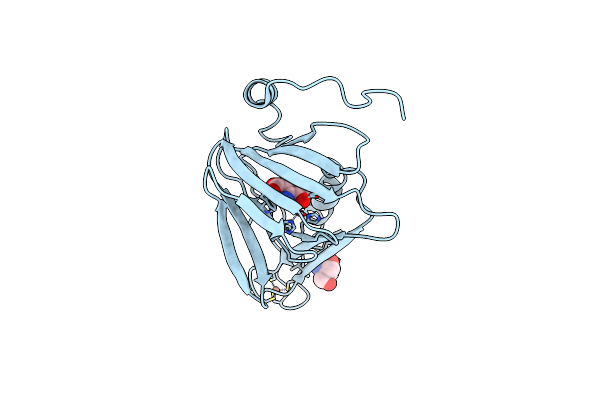
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - A Substrate Monodentately Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE2, 3HA |
 |
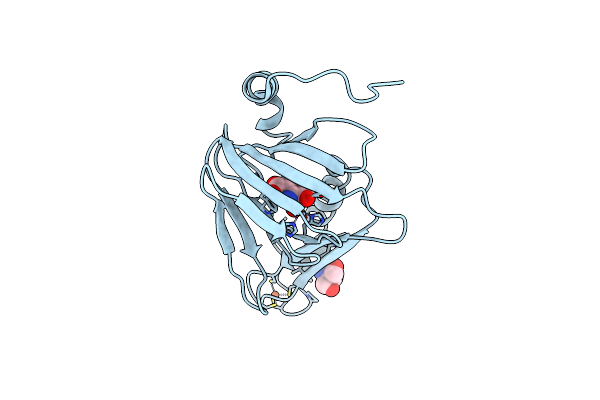
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - A Superoxo Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE, FE2, O, 3HA |
 |
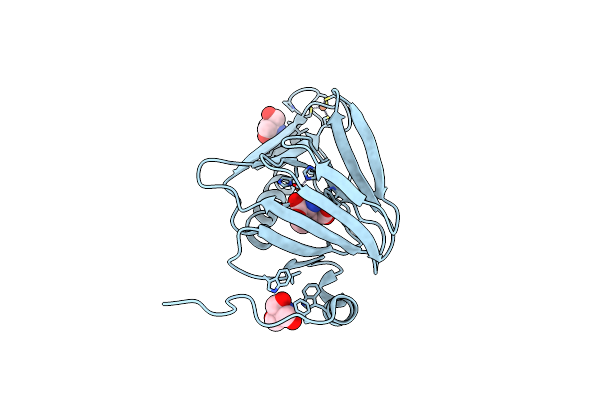
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - An Alkylperoxo Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: FE2, QXM, TRS |
 |
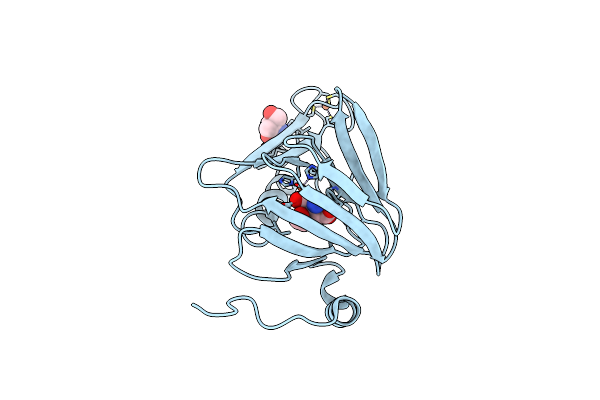
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - A Seven-Membered Lactone Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE2, FE, OH, EAY |
 |
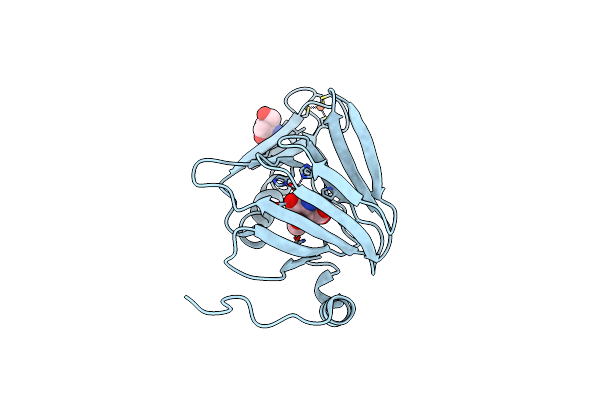
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - Enol Tautomers Of Acms Bidentately Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE2, 2FO |
 |
Observing A Ring-Cleaving Dioxygenase In Action Through A Crystalline Lens - An Enol Tautomer Of Acms Monodentately Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: TRS, FE2, 2FO |
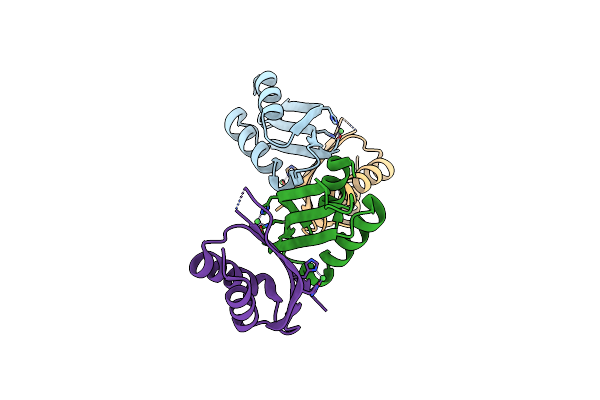 |
Metal-Bound C-Terminal Domain Of The Czcd Transporter From Cuprividus Metallidurans
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-06-24 Classification: TRANSPORT PROTEIN Ligands: NI |
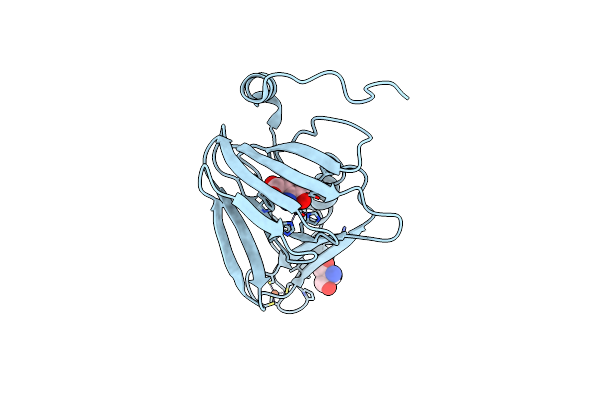 |
Probing Extradiol Dioxygenase Mechanism In Nad(+) Biosynthesis By Viewing Reaction Cycle Intermediates - A Substrate Bidentately Bound Structure
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE Ligands: 3HA, FE2, TRS |
 |
Crystal Structure Of 3-Hydroxyanthranilate-3,4-Dioxygenase I142P From Cupriavidus Metallidurans
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-06-06 Classification: OXIDOREDUCTASE Ligands: FE2, TRS |
 |
Crystal Structure Of 3-Hydroxyanthranilate-3,4-Dioxygenase I142P From Cupriavidus Metallidurans In Complex With 4-Cl-3-Haa
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-06-06 Classification: OXIDOREDUCTASE Ligands: FE2, 4AA, TRS |
 |
Crystal Structure Of 3-Hydroxyanthranilate-3,4-Dioxygenase I142P From Cupriavidus Metallidurans In Complex With 3-Haa
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2018-06-06 Classification: OXIDOREDUCTASE Ligands: FE2, 3HA, TRS |
 |
1.78 Angstrom Crystal Structure Of P97H 3-Hydroxyanthranilate-3,4-Dioxygenase From Cupriavidus Metallidurans
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
2.35 Angstrom Crystal Structure Of P97V 3-Hydroxyanthranilate-3,4-Dioxygenase From Cupriavidus Metallidurans
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE Ligands: FE2, TRS |
 |
2.72 Angstrom Crystal Structure Of P97A 3-Hydroxyanthranilate-3,4-Dioxygenase From Cupriavidus Metallidurans
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE Ligands: FE2, TRS |
 |
Crystal Structure Of The Transcription Regulator Pbrr691 From Ralstonia Metallidurans Ch34 In Complex With Lead(Ii)
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2016-12-28 Classification: TRANSCRIPTION Ligands: PB |
 |
Nmr Structure Of A 180 Residue Construct Encompassing The N-Terminal Metal-Binding Site And The Membrane Proximal Domain Of Silb From Cupriavidus Metallidurans Ch34
Organism: Cupriavidus metallidurans
Method: SOLUTION NMR Release Date: 2016-05-18 Classification: METAL BINDING PROTEIN Ligands: AG |

