Search Count: 15
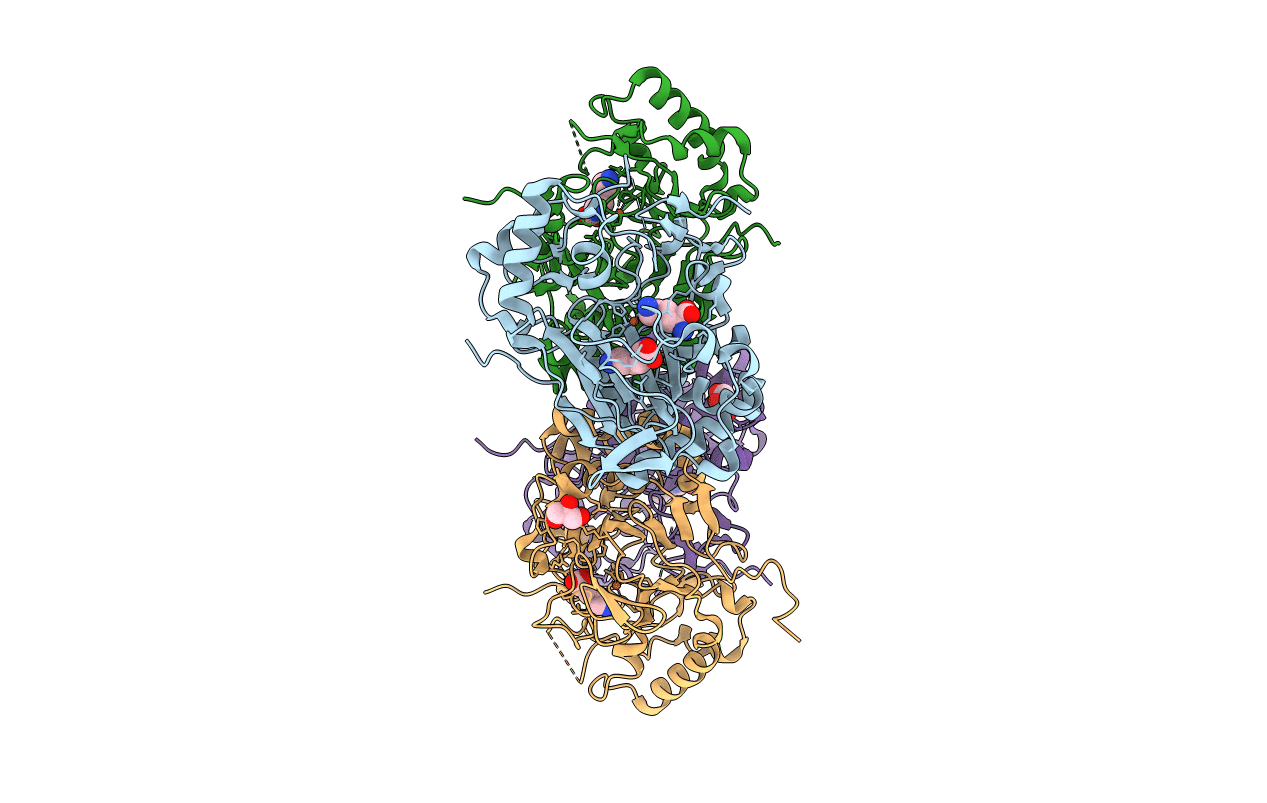 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Fe(Ii)/Lysine
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, LYS, GOL |
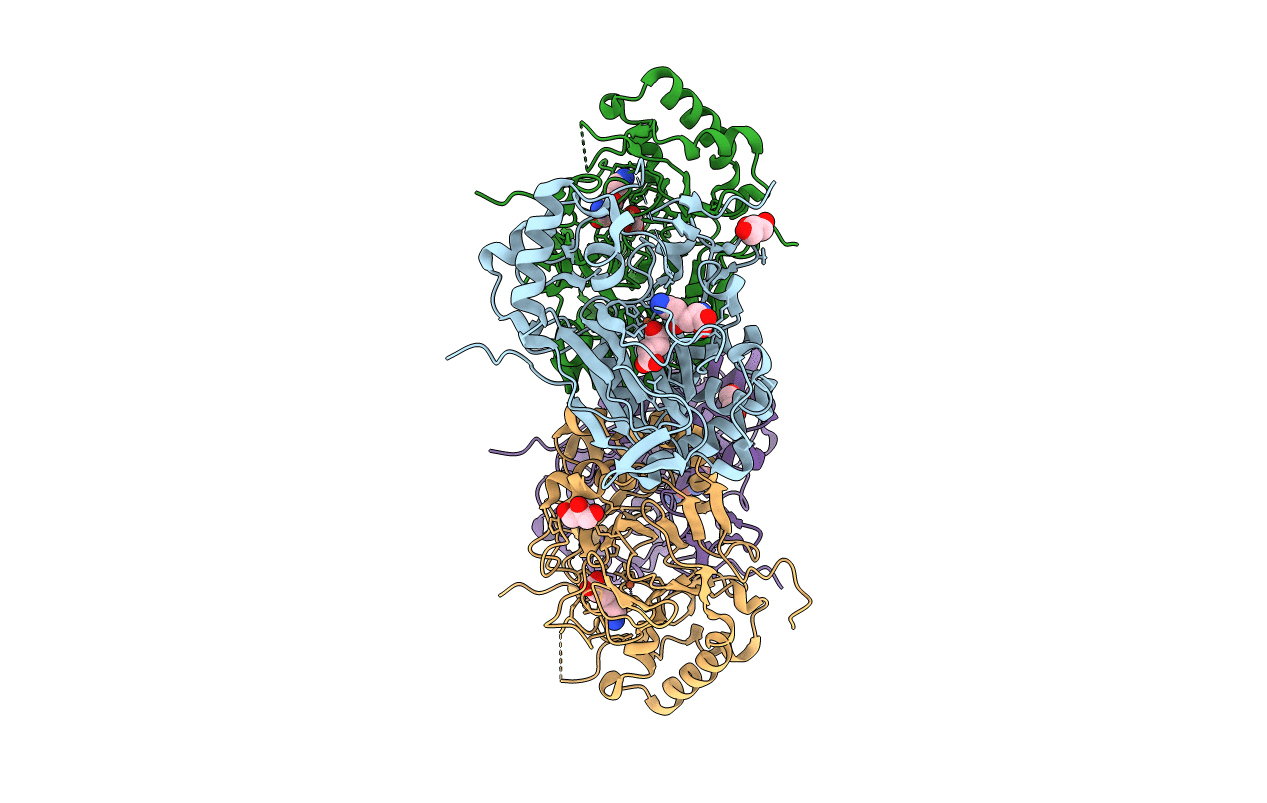 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Fe(Ii)/Succinate/(4R)-4-Hydroxy-L-Lysine
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, LYO, SIN, GOL |
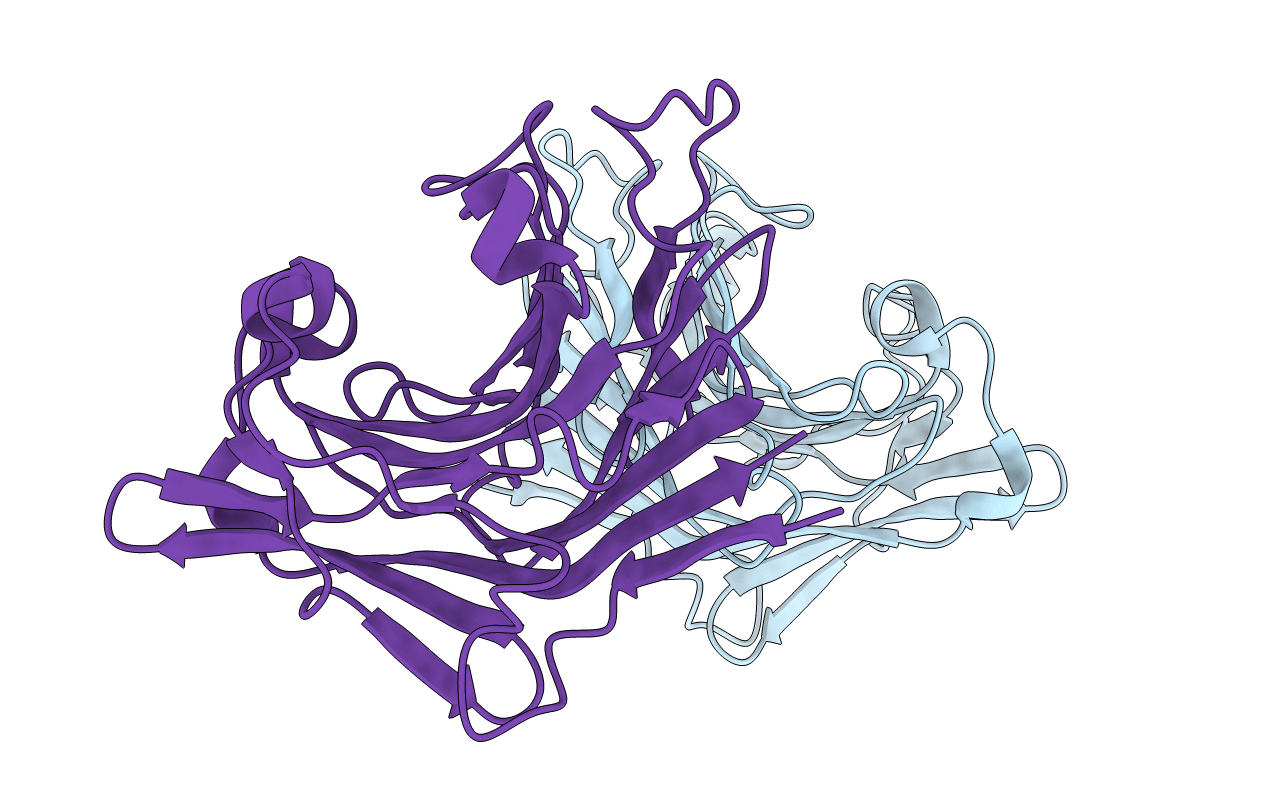 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-01-03 Classification: HYDROLASE |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: SO4, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y Mutant (Hyb-S4M94)
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: SO4, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, S112A/D370Y Mutant Complexed With 6-Aminohexanoate-Dimer
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
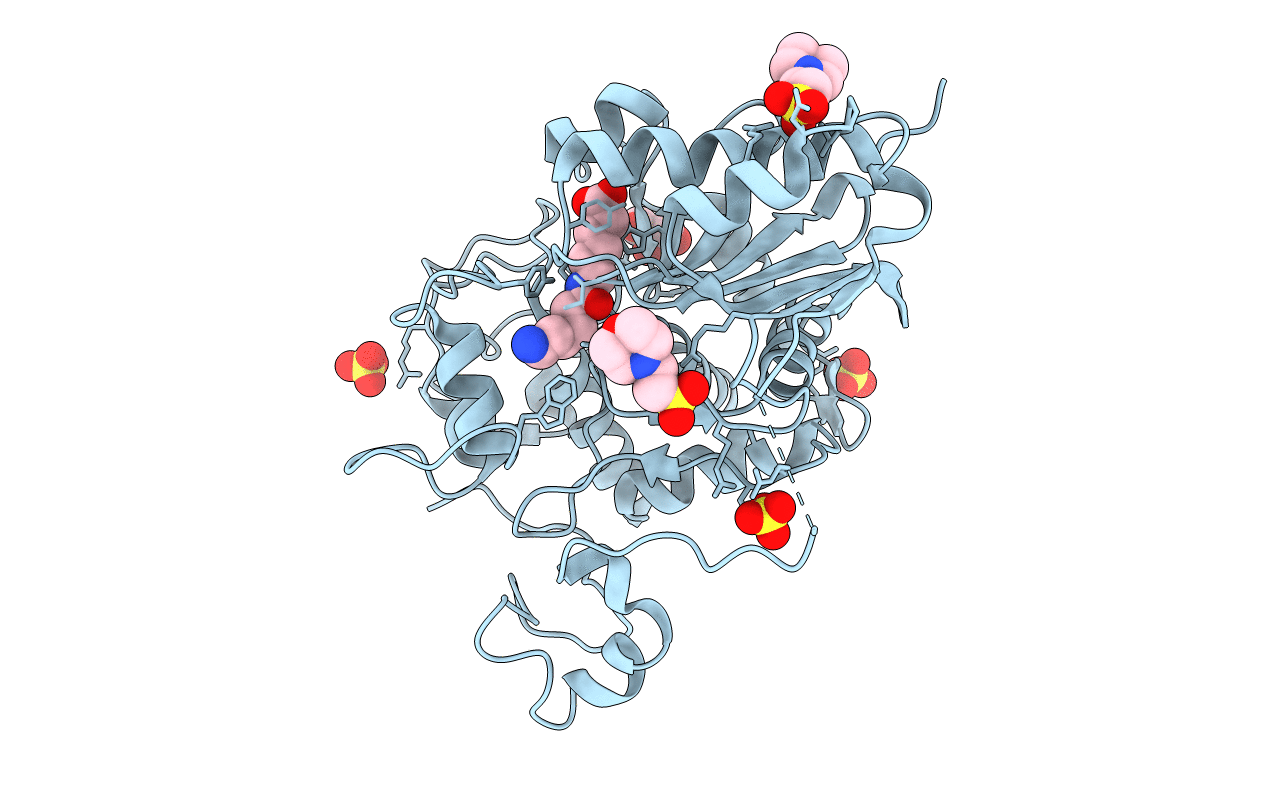 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y Mutant (Hyb-S4M94) With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, MES, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, S112A/G181D Mutant Complexed With 6-Aminohexanoate-Dimer
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N/D370Y Mutant With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N Mutant With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-01-09 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2006-02-21 Classification: HYDROLASE |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-09-20 Classification: HYDROLASE |
 |
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-09-16 Classification: HYDROLASE Ligands: ZN, EBP, EFS |
 |
Crystal Structure Of Phosphotriesterase Triple Mutant H254G/H257W/L303T Complexed With Diisopropylmethylphosphonate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-09-16 Classification: HYDROLASE Ligands: ZN, EBP, DII |

